To successfully develop from a single cell to a multicellular organism, the embryo must respond to mechanical and chemical signals that trigger its epithelial cells to divide, morph, and move. An additional constraint is that the cells must stay in contact on the apical surface to maintain the integrity of the epithelial tissue, which protects the developing internal organs of the embryo.
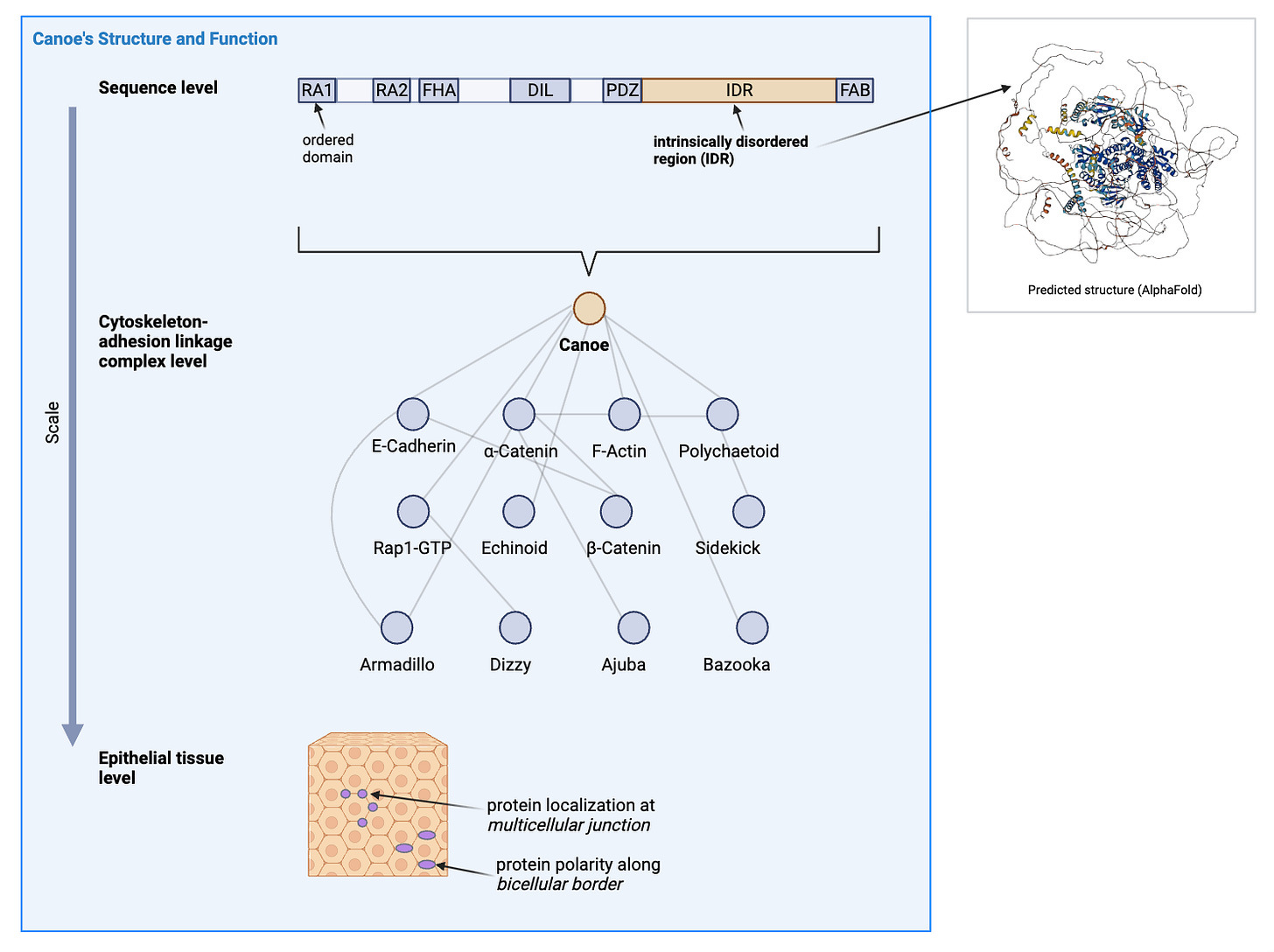
A network of scaffold proteins that link the force-generating cytoskeleton to the cell-cell adhesion junction proteins has been shown to be responsible for the balance between maintaining adhesion and enabling cell change. The multivalency (more than one binding interaction from each protein node) of the scaffold network is hypothesized to contribute to the observed robustness of the linkage in live organisms. In other words, we want to know which parts of the proteins are participating in the force-sensing and binding that brings the scaffold to support tension at the cell-cell interfaces effectively. Related questions that we have are where (relative to the cell-cell geometry) and when (in the developmental stages) are these proteins aggregating at the surface.
While we do know that in the big picture, each protein has differing essentiality to a viable or functional organism, we don’t know the relative importance of mechanical responses and biochemical interactions. A member of this scaffold network is the protein Canoe, which has been subject to domain function analysis for the last decade. The Canoe has a long intrinsically disordered region (IDR) that has been hypothesized to either bind or facilitate binding as a length between sites.
In this project with Prof. Peifer and Dr. Jensen, we focus on the IDR by using CRISPR to engineer drosophila melanogaster mutants that have portions of its IDR deleted or scrambled to investigate the effect of length and sequence. We then obtain the maternal-zygotic mutant embryos and use confocal microscopy to assess the state of the protein and the tissue. Our preliminary results would give data to assess theories about the specific functions of the IDR.
Afternotes
I have been procrastinating on an abstract for the current project and finally finished now that I have been in bed for flu for three days.
After reading my long-ass draft, Mark gave me valuable feedback: (1) write in full prose instead of bullet points of incomplete sentences, (2) don’t put any figures that an outsider cannot understand without you explaining to them more, (3) imitate what others do because the point is to just update the lab website bro.
The third point is good to know because I thought I had to do a journal style significance statement and think of all the wording from scratch, when really I just need to edit 50 words of the 300 words from other people on similar projects.
Implementing the first suggestion was still a recurrent challenge: how to complete a deliverable on time by working backwards and breaking down the tasks well from the time constraint; how to overcome mental temptation to run away from the outline and rewrite.
Fever made my head slower which was surprisingly a good thing because then I expected myself to do less things a day and also did not give in to the spiral of ideas when I started drafting. No doubt that the result now is still quite unclear, but at least I sent it.
I had a lot of fun revising the figure in BioRender, which is an online image editor that has a bunch of Creative Commons icons for biological entities. Back in May, Ellie introduced me to Monica Granados who works with Creative Commons. Dr. Granados told us about how BioRender is really great for many molecular biologists and it is a public good to have artists make new icons that the researchers need to represent their work in interpretable ways. I experienced this constraint of icon availability in that I couldn’t find any representing the adherens junction.
Dr. Granados also mentioned that there have been “jams” where artists and researchers come together for a few days and make new icons. That sounds really fun to me since I have been playing with diagrams ever since I started learning biology this year. I think the complexity and interrelations can really benefit from a preliminary logical organisation that a diagram in the form of hierarchies or edges or direct visual annotation can bring. I can see that biologists working with microscopy rely a lot on inherent information in visuals — relative intensity, geometric patterns, exceptions. Some of these nuances are extracted from the image just by the eye and are largely protected as an intuitive expertise, while other aspects have become quantified and automated, which is rare because often the samples are too few and the process of analysis is very hard to describe from those tacit experts — almost like biases.
In general, I want to know if there are other ways to use visual aid to better understand things together. I could start by constructing a personal syllabus on diagramming and interviewing a few experts as mentors or inspirations in this area, dead or alive.
A note on writing: It has been a rich week of posting daily even when I thought I had nothing to say about this theme (the study of living stuff). Some have been random and some have been convergent. I especially benefitted from giving definitions to terms I got from readings. I also learned that recounting dialogs are great ways to bring up confusing topics to other people for discussion. I will continue doing that in future interfacing with people through words. Now I will return to posting once a week here for fun reflection. I’m shifting writing outlet to specific individuals now!